Prof. Ravi Muddashetty

Profile: Ravi Muddashetty got his Ph.D. from the University of Muenster, Germany and pursued his major postdoctoral work at Emory University, Atlanta. Ravi’s lab is primarily interested in understanding the regulation of activity-mediated protein synthesis in neurons and how it affects the development and functioning of the nervous system. The lab is keen to elucidate the link between synaptic signalling, energy metabolism, activity-mediated protein synthesis and their integration in shaping synaptic function in health and diseases. In order to achieve this goal, the lab focuses on neurodevelopmental disorder Fragile X Syndrome and neurodegenerative disorder Alzheimer’s Disease.
For the Latest Lab News follow us on Twitter: @RaviMLab
Role of translation regulation in neurodevelopment:
Investigating the regulation of protein synthesis and its role in synaptic signaling is a primary research interest of our lab. Aberrant protein synthesis and dysregulated synaptic signaling are among the major underlying causes for neurodevelopmental disorders such as Fragile X syndrome (FXS). FXS is the major monogenetic form of Autism Spectrum Disorder (ASD) caused due to the absence of the RNA binding protein FMRP. We have identified that FMRP regulates multiple facets of protein synthesis beginning from ribosome biogenesis in the nucleus to activity mediated translation at the synapse. We are currently investigating how these versatile functions of FMRP contribute to neuronal development and plasticity using animal models and human stem cell-based models of FXS.
FMRP is also a member of the microRNA Induced Silencing Complex (miRISC) along with other RNA Binding Proteins (RBPs) such as MOV10 and GW182. Through their dynamic interactions with AGO2-mRNA-microRNA complex, these RBPs provide reversibility to miRISC function which is critical for translation regulation in neurons. We are interested in understanding the role of these RBPs in regulating neuronal development and plasticity, both individually and as a part of the miRISC machinery.
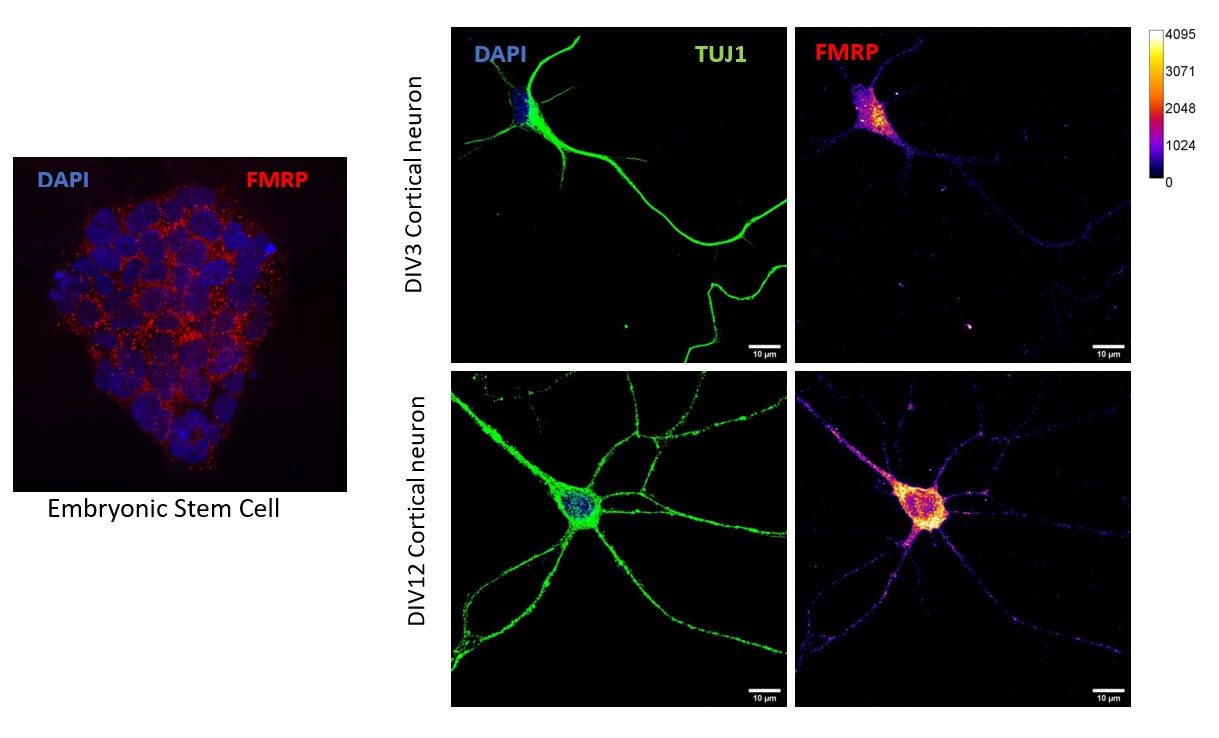
Role of translation regulation in neurodegeneration:
Our lab is interested in studying the defects of protein synthesis in the context of neurodegeneration, with a focus on Alzheimer’s disease (AD). We aim to understand how the dysregulation of protein synthesis could contribute to the synaptic dysfunction observed in AD. We focus on studying synaptic activity mediated protein synthesis and its defects in AD. To understand this, we study two major aspects – 1) the role of energy metabolism and 2) the role of calcium signaling, and how they are integrated to regulate synaptic/activity mediated protein synthesis. Exploring the link between energy metabolism and calcium homeostasis with synaptic protein synthesis is relevant in neurodegeneration as each of these aspects are individually shown to be affected. We use both human iPSC derived neurons and animal models of AD for our studies. In the case of iPSC derived neurons, we use iPSCs generated from familial AD patients, where the mutation has been corrected using CRISPR/Cas9 based gene editing. Similarly, we also use iPSCs from healthy individuals where the mutation has been knocked-in using CRISPR/Cas9 to generate the AD iPSC models. Apart from familial mutations, we also intend to make iPSCs from sporadic AD patients with known risk factors such as APOE4 to understand their contribution to AD pathology.

Translation regulation by epi-transcriptome:
Our lab is interested in understanding the role of non-coding RNAs like microRNA, snoRNA, rRNA and their interaction with RBPs in shaping neuronal function. We propose that the ribosome is not a mere chaperone, but a central platform for translation regulation through its interactions with the RBPs and the non-coding RNAs along their epi-transcriptomic modifications. Our lab is particularly interested in investigating the dynamics of translation regulation through RNA modifications like 2’O methylation and m6A modification in neurons. Our goal is to understand the molecular mechanism of translation regulation by epi-transcriptomic RNA modifications in healthy and diseased conditions.
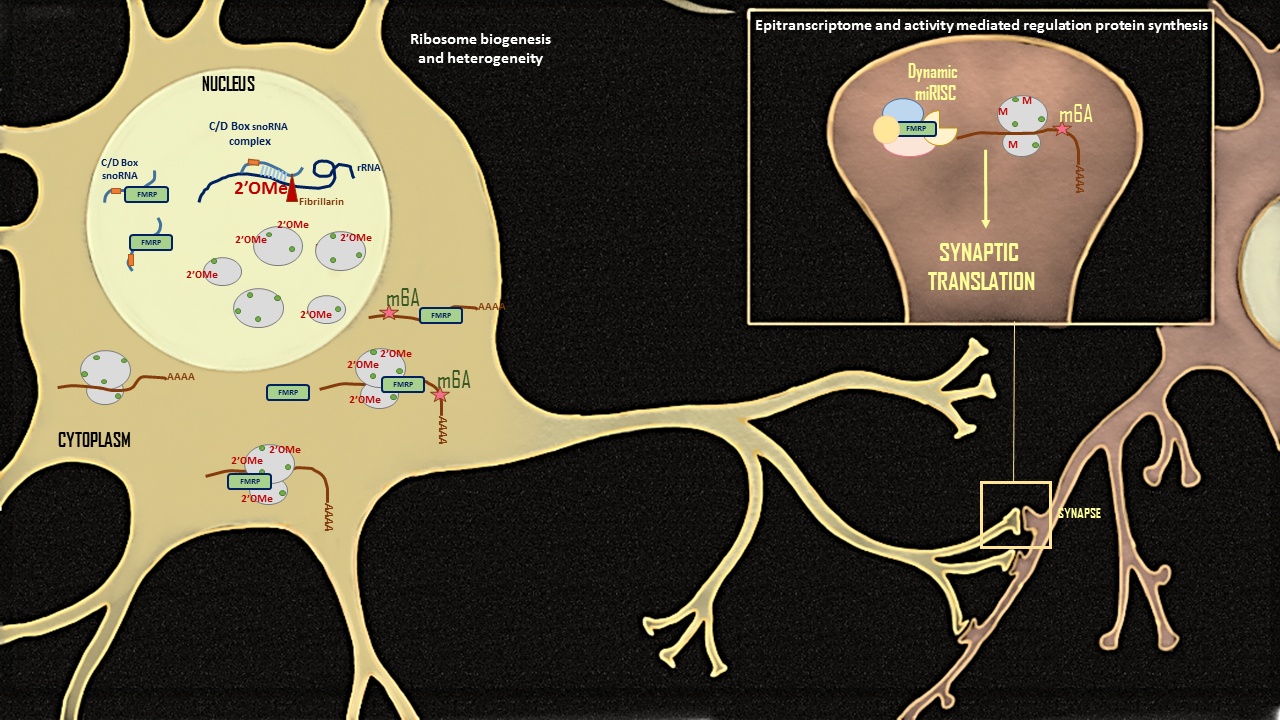
Present Lab Members
 Sarayu Ramakrishna – Postdoctoral Fellow
Sarayu Ramakrishna – Postdoctoral Fellow
Research Interest – The role of protein synthesis dysregulation in Alzheimer’s disease
 Michelle Ninochka D’Souza – PhD student
Michelle Ninochka D’Souza – PhD student
Research Interest – Structural contribution of Fragile X Mental Retardation Protein in neuronal translation regulation
 Bindushree K R – PhD student
Bindushree K R – PhD student
Research Interest – FMRP at the junction of Alzheimer’s and Autism Spectrum Disorder
 Nisa Shah – PhD student
Nisa Shah – PhD student
 Syed Wasifa Qadri – PhD student
Syed Wasifa Qadri – PhD student
 Shruti Pandey – PhD student
Shruti Pandey – PhD student
Former Lab Members
 Naveen Gowda
Naveen Gowda
St. John Medical Institute, Bangalore
 Bharti Nawalpuri
Bharti Nawalpuri
Cambridge University, UK
 Preeti M Kute – Post-doctoral Fellow
Preeti M Kute – Post-doctoral Fellow
University of Bergen, Norway
 Sreenath R – Post-doctoral Fellow
Sreenath R – Post-doctoral Fellow
Tata Institute of Fundamental Research
 Sudhriti G Dastidar
Sudhriti G Dastidar
NIH, USA
- Sarayu Ramakrishna, Bindushree K Radhakrishna, Ahamed P Kaladiyil1, Nisa Manzoor Shah, Nimisha Basavaraju,Kristine K Freude, Reddy Peera Kommadd, Ravi S Muddashetty. Distinct calcium sources regulate temporal profiles of NMDAR and mGluR-mediated protein synthesis.
Life Science Alliance. http://doi.org/10.26508/lsa.202402594 - Michelle Ninochka D’Souzal, Sarayu Ramakrishna, Bindushree K, Radhakrishna, Vishwaja Jhaveri, Sreenath Ravindran, Lahari Yeramala, Deepak Nair, Dasaradhi Palakodetil, Ravi S. Muddashetty. Function of FMRP Domains in Regulating Distinct Roles of Neuronal Protein Synthesis. Molecular Neurobiology. https://doi.org/10.1007/s12035-022-03049-1
- Naveen Kumar Chandappa Gowda, Bharti Nawalpuri, Sarayu Ramakrishna, Vishwaja Jhaveri & Ravi S. Muddashetty. NMDAR mediated dynamic changes in m6 A inversely correlates with neuronal translation. Scientific Reports (2022) 12:11317. https://doi.org/10.1038/s41598-022-14798-3
- Sarayu Ramakrishna, Vishwaja Jhaveri, Sabine Konings, Sumita Chakraborty, Bjorn Holst, Benjamin Schmid, Kristine Freude, Gunnar Gouras, Ravi Muddashetty . APOE4 affects basal and NMDAR mediated protein synthesis in neurons by perturbing calcium homeostasis. J Neurosci. 2021 Sep 2:JN-RM-0435-21. doi: 10.1523/JNEUROSCI.0435-21.2021. Online ahead of print.
- Bharti Nawalpuri and Ravi S Muddashetty. Distinct temporal expression of GW182 in neurons regulates dendritic arborization. J Cell Sci. 2021 Aug 15;134(16):jcs258465. doi: 10.1242/jcs.258465. Epub 2021 Aug 24.
- Ghosh Dastidar S, Das Sharma S, Chakraborty S, Chattarji S, Bhattacharya A, Muddashetty RS. Distinct regulation of bioenergetics and translation by group I mGluR and NMDAR. EMBO Rep. 2020 Jun 4;21(6):e48037. doi: 10.15252/embr.201948037. Epub 2020 Apr 29. PMID: 32351028; PMCID: PMC7271334.
- Nawalpuri B, Ravindran S, Muddashetty RS. The Role of Dynamic miRISC During Neuronal Development. Front Mol Biosci. 2020 Jan 31;7:8. doi: 10.3389/fmolb.2020.00008. PMID: 32118035; PMCID: PMC7025485.
- Kute PM, Ramakrishna S, Neelagandan N, Chattarji S, Muddashetty RS. NMDAR mediated translation at the synapse is regulated by MOV10 and FMRP. Mol Brain. 2019 Jul 10;12(1):65. doi: 10.1186/s13041-019-0473-0. PMID: 31291981; PMCID: PMC6617594.
- Kambali MY, Anshu K, Kutty BM, Muddashetty RS, Laxmi TR. Effect of early maternal separation stress on attention, spatial learning and social interaction behaviour. Exp Brain Res. 2019 Aug;237(8):1993-2010. doi: 10.1007/s00221-019-05567-2. Epub 2019 Jun 1. PMID: 31154461.
- Giri S, Purushottam M, Viswanath B, Muddashetty RS. Generation of a FMR1 homozygous knockout human embryonic stem cell line (WAe009-A-16) by CRISPR/Cas9 editing. Stem Cell Res. 2019 Aug;39:101494. doi: 10.1016/j.scr.2019.101494. Epub 2019 Jun 28. PMID: 31280136.
- Paul A, Nawalpuri B, Shah D, Sateesh S, Muddashetty RS, Clement JP. Differential Regulation of Syngap1 Translation by FMRP Modulates eEF2 Mediated Response on NMDAR Activity. Front Mol Neurosci. 2019 May 9;12:97. doi: 10.3389/fnmol.2019.00097. PMID: 31143100; PMCID: PMC6520660.
- Ramakrishna S, Muddashetty RS. Emerging Role of microRNAs in Dementia. J Mol Biol. 2019 Apr 19;431(9):1743-1762. doi: 10.1016/j.jmb.2019.01.046. Epub 2019 Feb 7. PMID: 30738891.
- Ravindran S, Nalavadi VC, Muddashetty RS. BDNF Induced Translation of Limk1 in Developing Neurons Regulates Dendrite Growth by Fine-Tuning Cofilin1 Activity. Front Mol Neurosci. 2019 Mar 20;12:64. doi: 10.3389/fnmol.2019.00064. PMID: 30949027; PMCID: PMC6436473.
- Schmid B, Prehn KR, Nimsanor N, Garcia BIA, Poulsen U, Jørring I, Rasmussen MA, Clausen C, Mau-Holzmann UA, Ramakrishna S, Muddashetty R, Steeg R, Bruce K, Mackintosh P, Ebneth A, Holst B, Cabrera-Socorro A. Generation of a set of isogenic, gene-edited iPSC lines homozygous for all main APOE variants and an APOE knock-out line. Stem Cell Res. 2019 Jan;34:101349. doi: 10.1016/j.scr.2018.11.010. Epub 2019 Jan 4. Erratum in: Stem Cell Res. 2020 Oct;48:102005. PMID: 30660866.
- Frederiksen HR, Holst B, Ramakrishna S, Muddashetty R, Schmid B, Freude K. Generation of two iPSC lines with either a heterozygous V717I or a heterozygous KM670/671NL mutation in the APP gene. Stem Cell Res. 2019 Jan;34:101368. doi: 10.1016/j.scr.2018.101368. Epub 2018 Dec 24. PMID: 30634129.
- D’Souza MN, Gowda NKC, Tiwari V, Babu RO, Anand P, Dastidar SG, Singh R, James OG, Selvaraj B, Pal R, Ramesh A, Chattarji S, Chandran S, Gulyani A, Palakodeti D, Muddashetty RS. FMRP Interacts with C/D Box snoRNA in the Nucleus and Regulates Ribosomal RNA Methylation. 2018 Nov 30;9:399-411. doi: 10.1016/j.isci.2018.11.007. Epub 2018 Nov 7. Erratum in: iScience. 2019 Feb 22;12:368. PMID: 30469012; PMCID: PMC6249352.
- Suhl JA, Muddashetty RS, Anderson BR, Ifrim MF, Visootsak J, Bassell GJ, Warren ST. A 3′ untranslated region variant in FMR1 eliminates neuronal activity-dependent translation of FMRP by disrupting binding of the RNA-binding protein HuR. Proc Natl Acad Sci U S A. 2015 Nov 24;112(47):E6553-61. doi: 10.1073/pnas.1514260112. Epub 2015 Nov 9. PMID: 26554012; PMCID: PMC4664359.
- Nalavadi VC, Muddashetty RS, Gross C, Bassell GJ. Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. J Neurosci. 2012 Feb 22;32(8):2582-7. doi: 10.1523/JNEUROSCI.5057-11.2012. PMID: 22357842; PMCID: PMC3427762.
- Muddashetty, Ravi S., Vijayalaxmi C. Nalavadi, and Gary J. Bassell. Fragile X Syndrome: A Disorder of Synaptic Protein Synthesis Dynamics. Journal of the Indian Institute of Science 92, no. 4 (December 28, 2012): 447–64.
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011 Jun 10;42(5):673-88. doi: 10.1016/j.molcel.2011.05.006. PMID: 21658607; PMCID: PMC3115785.
- Muddashetty RS, Keli? S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007 May 16;27(20):5338-48. doi: 10.1523/JNEUROSCI.0937-07.2007. PMID: 17507556; PMCID: PMC6672337.
- Khanam T, Muddashetty RS, Kahvejian A, Sonenberg N, Brosius J. Poly(A)-binding protein binds to A-rich sequences via RNA-binding domains 1+2 and 3+4. RNA Biol. 2006 Oct;3(4):170-7. doi: 10.4161/rna.3.4.4075. Epub 2006 Oct 26. PMID: 17387282.
- Kondrashov AV, Kiefmann M, Ebnet K, Khanam T, Muddashetty RS, Brosius J. Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP). J Mol Biol. 2005 Oct 14;353(1):88-103. doi: 10.1016/j.jmb.2005.07.049. PMID: 16154588.
- Muddashetty R, Khanam T, Kondrashov A, Bundman M, Iacoangeli A, Kremerskothen J, Duning K, Barnekow A, Hüttenhofer A, Tiedge H, Brosius J. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J Mol Biol. 2002 Aug 16;321(3):433-45. doi: 10.1016/s0022-2836(02)00655-1. PMID: 12162957.
Centre for Brain Research
Indian Institute of Science Campus
CV Raman Avenue
Bangalore 560012. India.
Email: ravimshetty[at]cbr-iisc.ac.in
Telephone: Office +91 80 2293 3588

