Prof. Bratati Kahali
DBT/Wellcome Trust India Alliance Intermediate Fellow
PhD: Bose Institute, India
Postdoc: University of Michigan Medical School, USA

Biosketch
Bratati Kahali, Ph.D., is an Associate Professor at the Centre for Brain Research. As a computational biologist focusing on human genetics and genomics, her research broadly examines the role of genetic variants in shaping our inherited traits and determining our predisposition to complex diseases, especially type 2 diabetes, dyslipidemia, cognition and neurodegeneration. Her lab members get trained and employ state-of-the-art approaches to study and analyze genetics and bioinformatics data, particularly in the Indian population, with special emphasis on whole genome and whole exome sequencing analyses. CBR is the coordinating centre for the national project, GenomeIndia and her lab is primarily involved in the whole genome sequencing data generation, processing and analysis.
Bratati earned her PhD at the Bose Institute (Kolkata, Jadavpur University, INDIA) in 2011 working at the Centre of Excellence in Bioinformatics. She was a postdoctoral fellow (2012-2016) at the University of Michigan Medical School (Ann Arbor, MI, USA), jointly affiliated with the Department of Internal Medicine (Gastroenterology division), and the Department of Computational Medicine and Bioinformatics. There, her research mainly focused on the identification of genetic loci for human obesity and non-alcoholic fatty liver disease, within the Genetic Investigation of Anthropometric Traits (GIANT) consortium and GOLD (Genetics of Lipid Diseases) consortium. In September 2016, she joined as a faculty member in Centre for Brain Research at the Indian Institute of Science.

Research
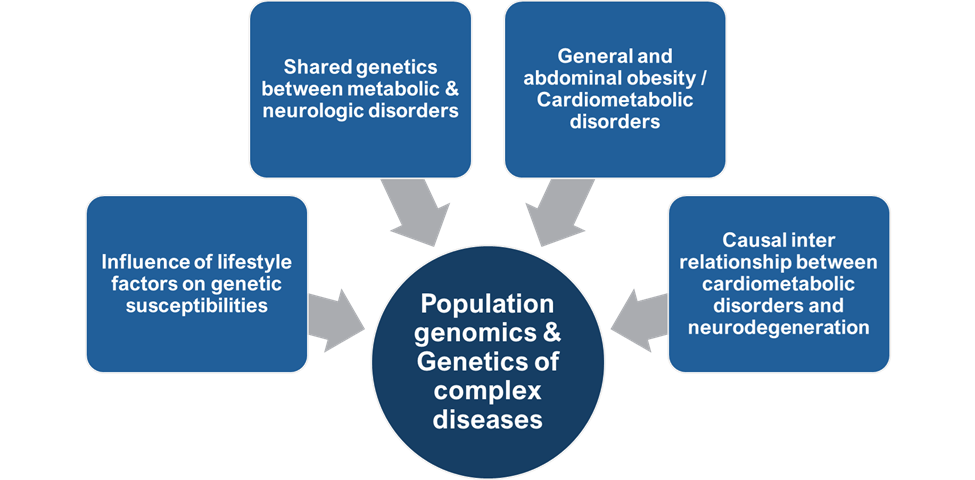
We use large-scale, high-throughput and next generation sequencing based genomic data to investigate how DNA sequence variants contribute to complex human diseases. The research in my group engages sophisticated statistical methodology and the use of high-performance computing resources for novel genetics analyses and methods development. The overarching goal of my laboratory is to develop and evaluate biological hypotheses for answering how human genetic variation can affect metabolic and neurodegenerative conditions – for example, why (a) humans have different genetic susceptibility to these complex diseases, and (b) find SNPs and structural variants that associate with the disease of interest, (c) unravel the physiological pathways relevant to disease being studied, (d) unravel causality and shared genetic basis of these disorders, (e) how gene-gene interactions contribute to shaping complex phenotypes and (f) how evolution have shaped these genomic characteristics; using integrative bioinformatics, statistical, and computational methods. We seek to integrate methylation, RNA expression and other -omics data in order to develop more powerful genomics-based predictors for such diseases.
Overall, my group’s research is aimed to better understand the shared genetic architecture between cardiometabolic disorders and neurodegenerative disorders, including genetic overlap between different disorders (i.e. pleiotropy) and genetic and phenotypic heterogeneity within disorders in Indian population. In addition, we aim to discern the causal interrelationships between cardiometabolic disorders and neurodegeneration.
Lab News
- Shreya Chakraborty (CBR-IMI) awarded the Fulbright- Nehru Fellowship, 2024.
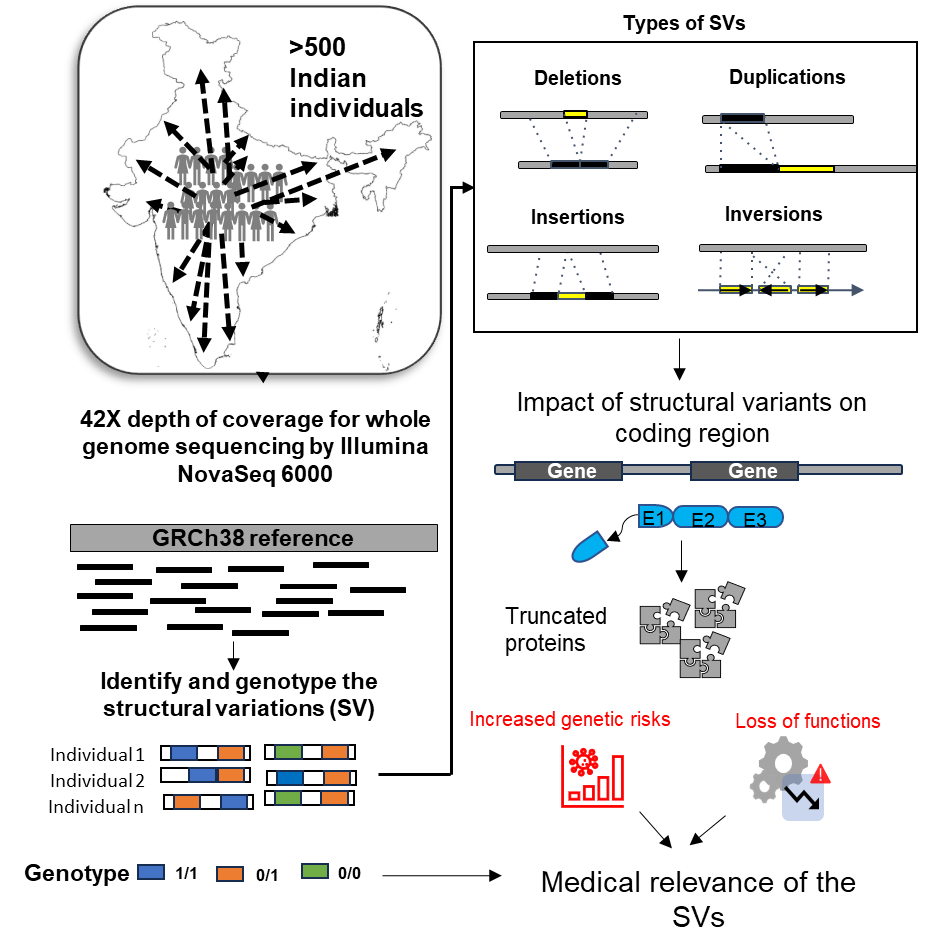
- Recent Publication: Landscape of genomic structural variations in Indian population-based cohorts: Deeper insights into their prevalence and clinical relevance. Krithika Subramanian, Mahek Chopra, Bratati Kahali. Human Genetics and Genomics Advances. doi: 10.1016/j.xhgg.2024.100285
- Dr. Bratati Kahali has been awarded the Wellcome Trust/ DBT India Alliance Intermediate Fellowship (January 2024).
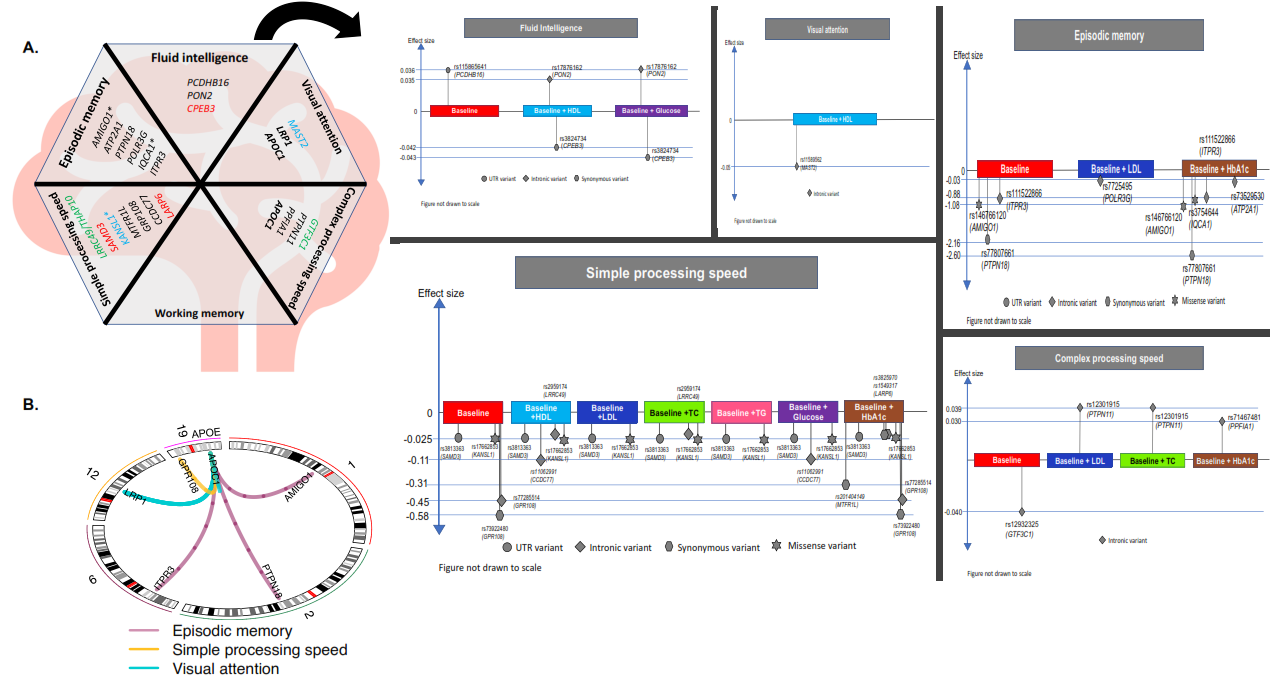
- Exome-wide analysis reveals role of LRP1 and additional novel loci in cognition. Shreya Chakraborty & Bratati Kahali. Human Genetics and Genomics Advances. https://doi.org/10.1016/j.xhgg.2023.100208
- Shreya Chakraborty (CBR-IMI) awarded the Prime Minister Research Fellowship (PMRF).
- Dr. Bratati Kahali has been awarded “National Supercomputing Mission 2020-2021”- Department of Science and Technology (DST) and Ministry of Electronics and Information Technology (MeITy), Government of India funding for “Population scale whole genome sequencing data analysis in Indians: Implications for uncovering the genetic architecture of cognitive changes associated with aging”.
- Allele Specific Variation at APOE Increases Non-alcoholic Fatty Liver Disease and Obesity but Decreases Risk of Alzheimer’s Disease and Myocardial Infarction.
Published by Dr. Bratati Kahali as lead author in Human Molecular Genetics. Accepted March 2021. - A Noncoding Variant Near PPP1R3B Promotes Liver Glycogen Storage and MetS, but Protects Against Myocardial Infarction. Kahali B, Chen Y, Feitosa MF, Bielak LF, O’Connell JR, Musani SK, Hegde Y, Chen Y, Stetson LC, Guo X, Fu YP, Smith AV, Ryan KA, Eiriksdottir G, Cohain AT, Allison M, Bakshi A, Bowden DW, Budoff MJ, Carr JJ, Carskadon S, Chen YI, Correa A, Crudup BF, Du X, Harris TB, Yang J, Kardia SLR, Launer LJ, Liu J, Mosley TH, Norris JM, Terry JG, Palanisamy N, Schadt EE, O’Donnell CJ, Yerges-Armstrong LM, Rotter JI, Wagenknecht LE, Handelman SK, Gudnason V, Province MA, Peyser PA, Halligan B, Palmer ND, Speliotes EK.J Clin Endocrinol Metab. 2021 Jan 23;106(2):372-387. doi: 10.1210/clinem/dgaa855.PMID: 33231259.
- Dr. Kahali was awarded the Ramalingaswami Fellowship 2016-2017 by Department of Biotechnology, Government of India.
- Dr. Kahali was awarded the Science and Engineering Research Board Early Career Fellow 2019 by Department of Science and Technology, Government of India.
- Lab member Mr. Abhay Gupta (undergraduate at IISc) was awarded the Travel fellowship for presenting and participating in Alzheimer’s Association International Conference, July 14-18, 2019 at Los Angeles, USA.


Present Members

Krithika Subramanian
Ph.D Student
About
Currently working on

Shreya Chakraborty
Ph.D Student
About
Currently working on

Jani Siddhi Pushkarbhai
About
M.Sc. Bioinformatics, Gujarat University, Ahmedabad, Gujarat
Currently working on

Tanmay Panigrahi
Ph.D Student
About
Currently working on

Sauma Suvra Majumdar
PhD student (IMI, IISc)
About
BTech-MTech dual degree in Biotechnology, National Institute of Technology, Durgapur
Currently working on
Identification and characterization of structural and copy number genetic variations in the human X and Y chromosomes and their polygenic effect on neurodegeneration

Samarpita Saha
Project Assistant

Yuvraj Sankhla
Project Intern
Former Members
Raghvendra Agrawal (Data Analyst)
Rupanwita Majumder (Data Analyst)
Akkshaya R (Data Analyst)
Aditya Yinaganti (Project Associate)
Sree Vishmaya V (Project Associate)
Mehak Chopra (Project Associate)
Shukrruthi K Iyengar (Project Intern)
Gowtham Manivel, PhD (Post-Doctoral Fellow)
Aarti Rana, PhD (Post-Doctoral Fellow)
Rajesh , PhD (Post-Doctoral Fellow)
Abhishek Panda (Project Associate)
Anupriya Sadhasivam (Project Assistant)
Swetaleena Satpathy (Data Analyst)
Sheldon D’Silva (Junior Research Fellow)
Shreya Jha (Project Assistant)
Abhay Gupta (Project Intern)
Shraddha Barke
Greeshma Thulasi
Rajesh Kumar Maurya
Sumanth Gowda
Krithika Subramanian, Mehak Chopra,1 and Bratati Kahali. Landscape of genomic structural variations in Indian population-based cohorts: Deeper insights into their prevalence and clinical relevance. Human Genetics and Genomics Advances. March 23, 2024, DOI:https://doi.org/10.1016/j.xhgg.2024.100285
https://www.cell.com/hgg-advances/fulltext/S2666-2477(24)00024-1
Chen Y, Du X, Kuppa A, Feitosa MF, Bielak LF, O’Connell JR, Musani SK, Guo X, Kahali B, Chen VL, Smith AV, Ryan KA, Eirksdottir G, Allison MA, Bowden DW, Budoff MJ, Carr JJ, Chen YI, Taylor KD, Oliveri A, Correa A, Crudup BF, Kardia SLR, Mosley TH Jr, Norris JM, Terry JG, Rotter JI, Wagenknecht LE, Halligan BD, Young KA, Hokanson JE, Washko GR, Gudnason V, Province MA, Peyser PA, Palmer ND, Speliotes EK.
Nat Genet. 2023 Oct;55(10):1640-1650. doi: 10.1038/s41588-023-01497-6. Epub 2023 Sep 14.
Genome-wide association meta-analysis identifies 17 loci associated with nonalcoholic fatty liver disease.
Kahali B, Balakrishnan A, Dhanavanthri Muralidhara S, Muniz-Terrera G, Ritchie K; SANSCOG Study Team; Ravindranath V. Alzheimers Dement. 2023 Jun;19(6):2433-2442. doi: 10.1002/alz.12572. Epub 2022 Dec 14.
COGNITO (Computerized assessment of adult information processing): Normative scores for a rural Indian population from the SANSCOG study.
Ravindranath V; SANSCOG Study Team. Alzheimers Dement. 2023 Jun;19(6):2450-2459. doi: 10.1002/alz.12722. Epub 2022 Dec 28. Bratati Kahali is contributing member in the SANSCOG study team.
Srinivaspura Aging, Neuro Senescence and COGnition (SANSCOG) study: Study protocol
Shreya Chakraborty & Bratati Kahali. Exome-wide analysis reveals role of LRP1 and additional novel loci in cognition. Human Genetics and Genomics Advances. https://doi.org/10.1016/j.xhgg.2023.100208
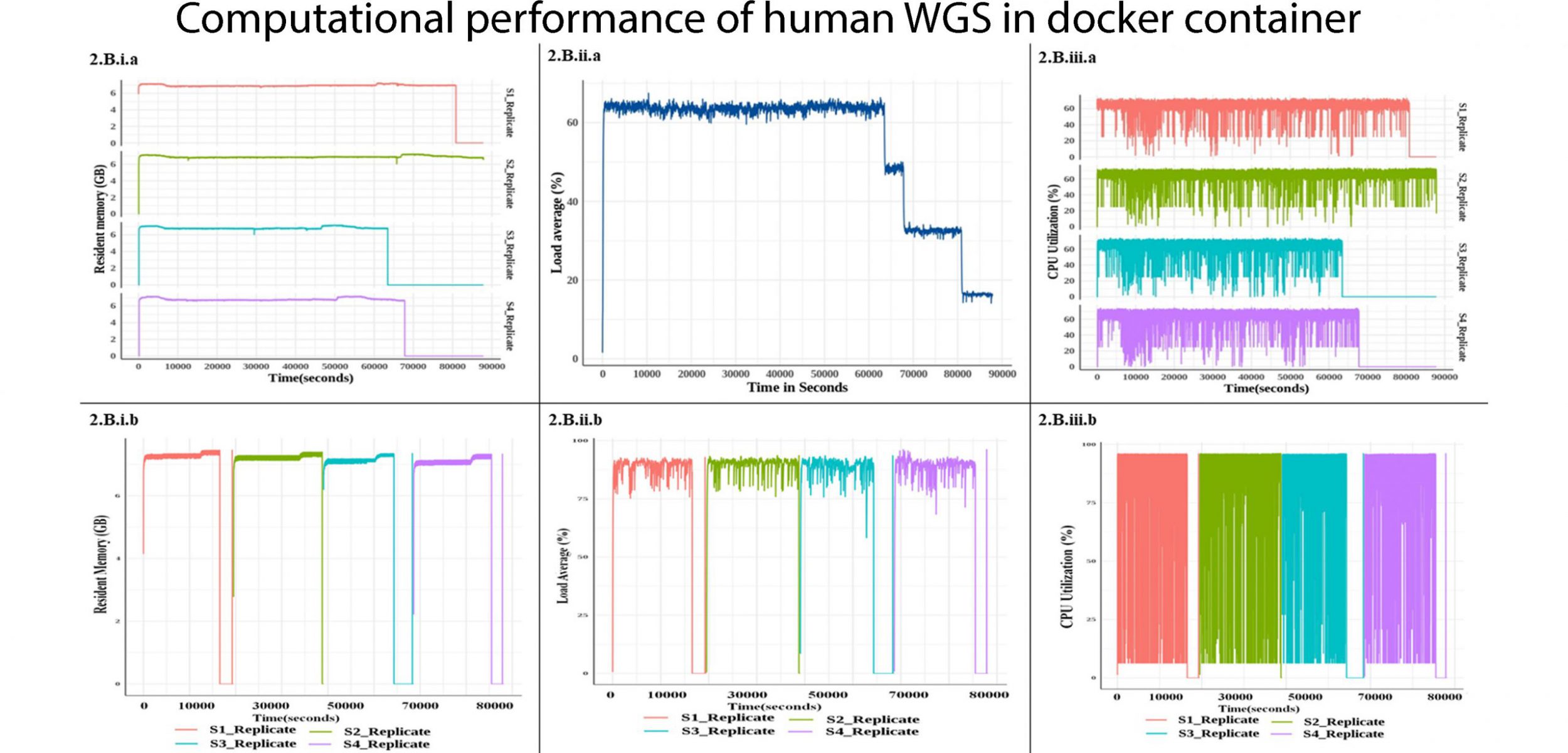
Abhishek Panda, Krithika Subramanian, Bratati Kahali. Implementation of human whole genome sequencing data analysis: A containerized framework for sustained and enhanced throughput. Elsevier Ltd. 30 July 2021. https://doi.org/10.1016/j.imu.2021.100684
Implementation of human whole genome sequencing data analysis: A containerized framework for sustained and enhanced throughput – ScienceDirect
Published by Dr. Bratati Kahali as lead author in Human Molecular Genetics. Allele Specific Variation at APOE Increases Non-alcoholic Fatty Liver Disease and Obesity but Decreases Risk of Alzheimer’s Disease and Myocardial Infarction. Accepted March 2021.
Kahali B, Chen Y, Feitosa MF, Bielak LF, O’Connell JR, Musani SK, Hegde Y, Chen Y, Stetson LC, Guo X, Fu YP, Smith AV, Ryan KA, Eiriksdottir G, Cohain AT, Allison M, Bakshi A, Bowden DW, Budoff MJ, Carr JJ, Carskadon S, Chen YI, Correa A, Crudup BF, Du X, Harris TB, Yang J, Kardia SLR, Launer LJ, Liu J, Mosley TH, Norris JM, Terry JG, Palanisamy N, Schadt EE, O’Donnell CJ, Yerges-Armstrong LM, Rotter JI, Wagenknecht LE, Handelman SK, Gudnason V, Province MA, Peyser PA, Halligan B, Palmer ND, Speliotes EK. A Noncoding Variant Near PPP1R3B Promotes Liver Glycogen Storage and MetS, but Protects Against Myocardial Infarction. J Clin Endocrinol Metab. 2021 Jan 23;106(2):372-387. doi: 10.1210/clinem/dgaa855.PMID: 33231259.
Gupta A,Bratati Kahali*, Machine learning-based cognitive impairment classification with optimal combination of neuropsychological tests. Alzheimers Dement (N Y), 2020 Jul 19;6(1):e12049. doi: 10.1002/trc2.12049. eCollection 2020. PMID: 32699817
Haycock P, Burgess S,…., Bratati Kahali*, …,et al. The association between genetically elevated telomere length and risk of cancer and non- neoplastic diseases. JAMA Oncology, Feb, 2017.
Marouli E, Graff M,…, Bratati Kahali*, …,et al. Rare and low-frequency coding variants alter human adult height. paper from GIANT consortium. Nature, 542(7640):186-190, 2017.
Adam E Locke*, Bratati Kahali*, Sonja I Berndt*, Anne E Justice*, Tune H Pers*,……………….,Gonçalo R Abecasis, Lude Franke, Timothy M Frayling, Mark I McCarthy, Peter M Visscher, André Scherag, Cristen J Willer, Michael Boehnke, Karen L Mohlke, Cecilia M Lindgren, Jacques S Beckmann, Inês Barroso, Kari E North§, Erik Ingelsson§, Joel N Hirschhorn§, Ruth JF Loos§, Elizabeth K Speliotes§ Genetic studies of body mass index yield new insights for obesity biology. Nature 518(7538), 197-206, 2015.* Equal contribution
Dmitry Shungin, Thomas W Winkler, Damien C Croteau-Chonka, Teresa Ferreira, …, Bratati Kahali, ….., Cristen J Willer, Peter M Visscher, Jian Yang, Joel N Hirschhorn, M Carola Zillikens, Mark I McCarthy, Elizabeth K Speliotes, Kari E North, Caroline S Fox, Inês Barroso, Paul W Frank, Erik Ingelsson, Iris M Heid4, Ruth JF Loos, L Adrienne Cupples, Andrew P Morris, Cecilia M Lindgren, Karen L Mohlke. New genetic loci link adipose and insulin biology to body fat distribution. Nature 518(7538), 187-196, 2015.
Bratati Kahali, Liu YL, Daly AK, Day CP, Anstee QM, Speliotes EK. TM6SF2: Catch-22 in the Fight Against Nonalcoholic Fatty Liver Disease and Cardiovascular Disease?Gastroenterology 148(4), 679-684, 2015.
Matthew R. Robinson, Gibran Hemani, ……, Bratati Kahali,……Elizabeth K. Speliotes, Michael E. Goddard, Jian Yang, Peter M. Visscher. Population genetic differentiation of height and body mass index across Europe. Nature Genetics 47(11),1357-1362, 2015.
Sara Hägg, Andrea Ganna, Tonu Esko, Sander W van der Laan, Adam E Locke, Bratati Kahali, Sonja I Berndt, Anne E Justice,, Tune H Pers, Kari E North, Ruth JF Loos, Elizabeth K Speliotes, Timothy M Frayling, Peter M Visscher, David P Strachan, Joel N Hirschhorn, Yudi Pawitan, Erik Ingelsson. Gene-based meta-analysis of genome-wide association studies implicates new gene regions involved in obesity. Human Molecular Genetics 24(23), 6849-6860, 2015.
Nicholette D Palmer, Solomon K Musani, Laura M Yerges, Mary F Feitosa, Lawrence F Bielak, Ruben Hernaez, Bratati Kahali, J Jeffrey Carr, Tamara B Harris, Min A Jhun, Sharon LR Kardia, Carl D Langefeld, Thomas H Mosley Jr, Jill M Norris, Albert V Smith, Herman A Taylor, Lynne E Wagenknecht, Jiankang Liu, Ingrid B Borecki, Patricia A Peyser, Elizabeth K Speliotes. Characterization of European –ancestry NAFLD Associated Variants in Individuals of African and Hispanic Descent. Hepatology 58(3), 966-975, 2013.
Aaron P. Thrift, Jian Gong, Ulrike Peters, Jenny Chang-Claude, Anja Rudolph, Martha L. Slattery, Andrew T. Chan, Adam E. Locke, Bratati Kahali, …, Michael Hoffmeister, Hermann Brenner, Emily White, Li Hsu, Peter T. Campbell. Mendelian randomization study of body mass index and colorectal cancer risk. Cancer Epidemiology, Biomarkers & Prevention 24(7), 1024-1031, 2015.
Bratati Kahali, Brian Halligan, Elizabeth K Speliotes. Insights from genome wide association analyses of nonalcoholic fatty liver disease. Seminars in Liver Disease 35(4), 375-391, 2015.
Bratati Kahali, Tapash Chandra Ghosh. Disorderness in Escherichia coli proteome: perception of folding fidelity and protein-protein interactions. J Biomol Struct Dyn 31(5), 472-476, 2013.
Bratati Kahali *, Shandar Ahmad, Tapash Chandra Ghosh*. Selective constraints in yeast genes with differential expressivity: Codon pair usage and mRNA stability perspectives. Gene 481, 76-82, 2011.*Joint corresponding author
Sandip Chakraborty, Bratati Kahali *, Tapash Chandra Ghosh*. Protein complex forming ability is favored over the features of interacting partners in determining the evolutionary rates of proteins in the yeast protein-protein interaction networks. BMC Syst Biol 4:155, 2010.*Joint corresponding author
Singh YH, Andrabi M, Bratati Kahali, Ghosh TC, Mizuguchi K, Kochetov AV, Ahmad S. On nucleotide solvent accessibility in RNA structure. Gene 463(1-2), 41-48, 2010.
Bratati Kahali, Shandar Ahmad and Tapash Chandra Ghosh. Exploring the evolutionary rate differences of party hub and date hub proteins in Saccharomyces cerevisiae protein-protein interaction network. Gene 429(1-2), 18-22, 2009.
Baisali Manna, Tanusree Bhattacharya, Bratati Kahali, Tapash Chandra Ghosh. Evolutionary constraints on hub and non-hub proteins in human protein interaction network: insight from protein connectivity and intrinsic disorder. Gene 434(1-2), 50-55, 2009.
Bratati Kahali, Surajit Basak and Tapash Chandra Ghosh. Delving deeper into the unexpected correlation between gene expressivity and codon usage bias of Escherichia coli genome. J Biomol Struct Dyn. 25, 6556-6561, 2008.
Bratati Kahali, Surajit Basak and Tapash Chandra Ghosh. Reinvestigating the codon and amino acid usage of S. cerevisiae genome: A new insight from protein secondary structure analysis. Biochemical and Biophysical Research Communications 354, 693-699, 2007.
Book Chapter(s):
Bratati Kahali and Elizabeth K. Speliotes. Genetic Pleiotropies of Obesity.
The Genetics of Obesity, Edited by Dr. Struan Grant.
SpringerLink ISBN: 978-1-4614-8641-1
Sandip Chakraborty, Soumita Podder, Bratati Kahali, Tina Begum, Kamalika Sen, and Tapash Chandra Ghosh. (All authors contributed equally). Insights into Eukaryotic Interacting Protein Evolution.
Evolutionary Biology – Concepts, Biodiversity, Macroevolution and Genome Evolution.
Springer-Verlag Berlin Heidelberg. Pontarotti, Pierre (Ed.) 1st Edition., 2011, XV, ISBN 978-3-642-20762-4.

