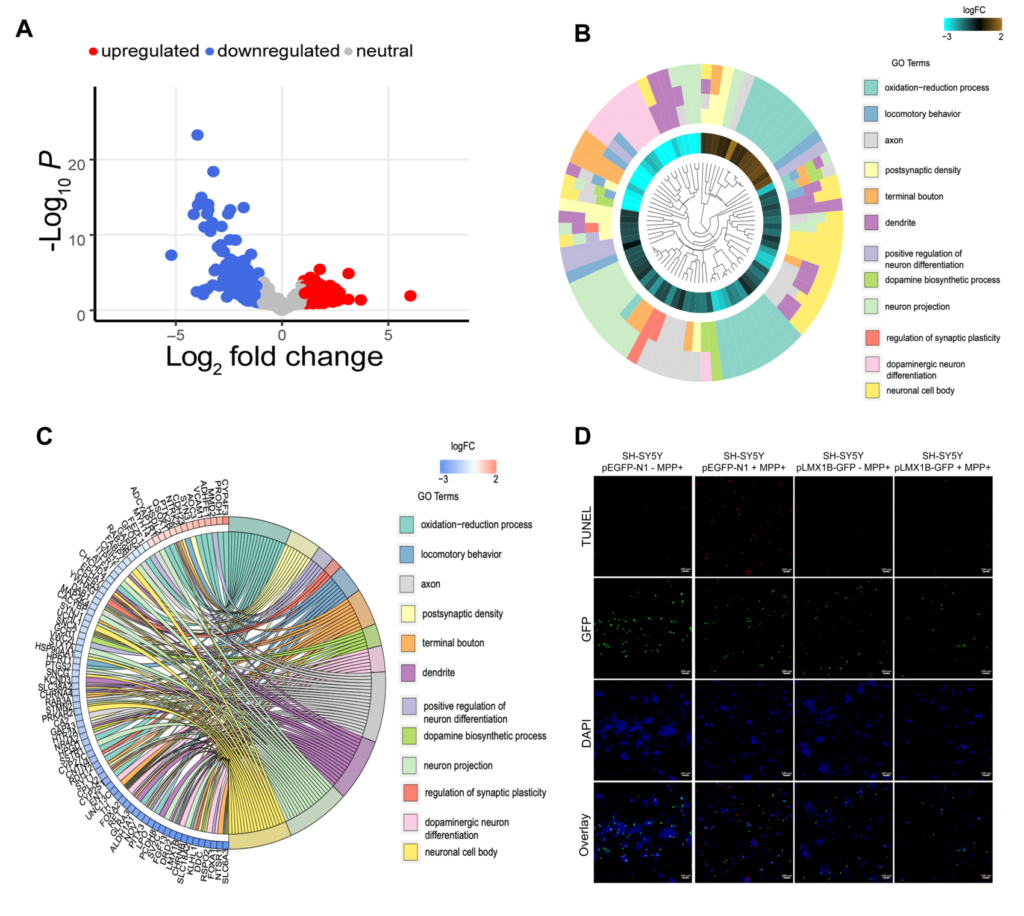
Parkinson’s disease (PD) is a debilitating neurodegenerative movement disorder. The cardinal clinical symptoms of PD include rigidity, bradykinesia and resting tremor. PD is characterized by the degeneration of dopaminergic neurons in the substantia nigra pars compacta (SNpc). However, the exact cause underlying loss of dopaminergic neurons in SNPc in PD remains poorly understood. Therefore, understanding the mechanisms that lead to neurodegeneration of dopaminergic neurons in SNpc is vital. We performed transcriptome analysis using RNA sequencing on substantia nigra pars compacta (SNpc) from mice after acute and chronic 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) treatment and PD patients. We found that acute and chronic exposure to MPTP resulted in decreased expression of genes involved in sodium channel regulation. Dopamine biosynthesis and synaptic vesicle recycling pathways were downregulated in PD patients and after chronic MPTP treatment in mice. Genes essential for midbrain development and determination of dopaminergic phenotype such as, LMX1B, LMX1A, GAP43, SNCA, PBX1, and GRB10 were downregulated in PD patients. Further, overexpression of LMX1B rescued MPP+ induced death in SH-SY5Y neurons. We conclude that the downregulation of gene ensemble involved in development and differentiation of dopaminergic neurons indicate their potential involvement in pathogenesis and progression of PD in humans.
Last Updated on March 14, 2024
References :
Verma A, Kommaddi RP, Gnanabharathi B, Hirsch EC, Ravindranath V. Genes critical for development and differentiation of dopaminergic neurons are downregulated in Parkinson’s disease. J Neural Transm (Vienna). 2023 Apr;130(4):495-512. PMID: 36820885.
Verma A, Kommaddi RP, Gnanabharathi B, Hirsch EC, Ravindranath V. Genes critical for development and differentiation of dopaminergic neurons are downregulated in Parkinson’s disease. J Neural Transm (Vienna). 2023 Apr;130(4):495-512. PMID: 36820885.
![]()
